GEODEMO_4b
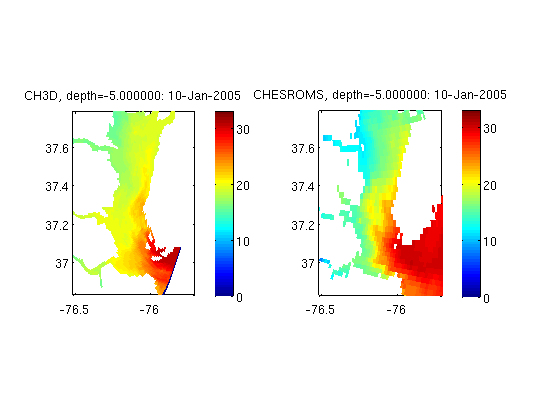
Compare horizontal slices from two different CF compliant structured grid models (CH3D and ROMS) at a particular time step and depth, using geosubset to subset the data
Contents
CH3D
url{1}='http://testbedapps.sura.org/thredds/dodsC/estuarine_hypoxia/ch3d/agg.nc';
var{1}='salinity';
titl{1}='CH3D';
ROMS
url{2}='http://testbedapps.sura.org/thredds/dodsC/estuarine_hypoxia/chesroms_1tdo/agg.nc';
var{2}='salt';
titl{2}='CHESROMS';
Create geosubset object
dat=[2005 1 10 0 0 0]; % Jan 10, 2005 00:00 UTC depth=-5; % horizontal slice 5 m from surface ax=[ -76.5220 -75.7105 36.8248 37.7850]; %lon/lat range cax=[0 33]; %color range lat_mid=38; % for scaling plots s.time=dat; s.lon=ax(1:2); s.lat=ax(3:4);
Perform analysis without using dataset dependant code
Access datasets, subset data, interpolate data to a constant z, plot results at z depth
figure; for i=1:length(url); nc{i}=ncgeodataset(url{i}); % create a salinity geovariable object. No data read yet. svar{i}=geovariable(nc{i},var{i}); disp(['reading data from ' titl{i} '...']) % using geosubset here, which allows for subsetting and striding, reading % multiple time steps, only certain zlevels and more. sub{i}=svar{i}.geosubset(s); sz{i}=zsliceg(squeeze(sub{i}.data),squeeze(sub{i}.grid.z),depth); a{i}=subplot(1,length(url),i); pcolorjw(sub{i}.grid.lon,sub{i}.grid.lat,double(sz{i}));colorbar axis(ax); caxis(cax); title(sprintf('%s, depth=%f: %s',titl{i},depth,datestr(sub{1}.grid.time))); set (a{i}, 'DataAspectRatio', [1 cos(lat_mid*pi/180) 1000] ); end
reading data from CH3D... reading data from CHESROMS...